Evolution and adaptation are, by definition, multigenerational processes. An exciting recent insight has been the extent to which opposing forces of spatially diversifying selection and homogenizing gene flow can result in local adaptation at small spatial scales relative to average dispersal distances (Richardson et al. 2014). Multiple mechanisms that can account for this result in theory, such as habitat choice by dispersal propagules, potentially explain patterns that have seemed surprising based on simple analytical theory. In this blog post we focus on an underappreciated nonequilibrium process that has been in plain sight all along, potentially confounding local adaptation inferences, but has been considered trivial.
What importance does selection have, you might wonder, if high gene flow prevents local adaptation? When Hermon Bumpus revived 72 out of 136 house sparrows he found downed by a violent winter storm in 1898, the survivors had significant differences in weight and morphology that he and later analysts attributed to natural selection (Bumpus 1899; Grant 1972). Episodic selection like this can be interesting and informative to study, but probably changes little in the population’s evolutionary trajectory.
In contrast, marine benthic species and many terrestrial plants are sedentary as adults, highly fecund, and profoundly vulnerable to high mortality in their earliest life stages during dispersal and soon after settlement. When each female spawns tens of millions of eggs and larval dispersal is broad there is much more potential for within-generation responses to selection across habitat heterogeneities than if only a few embryos are produced. For species with this life history it is not known how much early mortality is effectively random (from a mix of factors) versus selective across biophysical environmental gradients. However, a few classic examples have demonstrated “adaptive” differentiation on such a small spatial scale, recurring each life cycle, that no other mechanism besides within-generation selection is possible to explain the differentiation. Terms used in the literature to describe this process include ‘phenotype-environment mismatch’, and ‘balanced polymorphism’ for the end result. Here, to put emphasis on the recurrent selection regenerating differentiation in each cohort, we use “cohort adaptation” to contrast with local adaptation (Fig. 1).
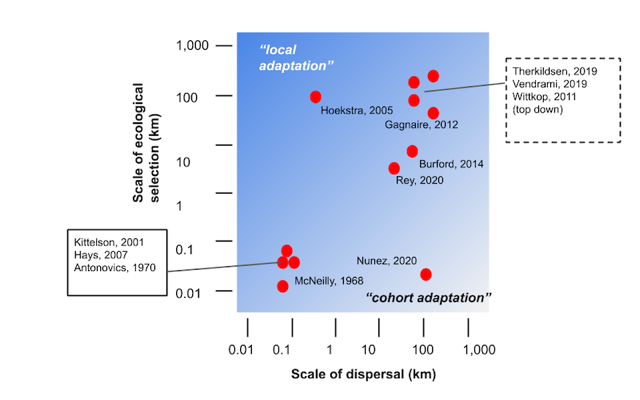
Fig 1: Local adaptation vs. cohort adaptation. Examples of adaptive differentiation plotted relative to the spatial scale of the selection gradient vs average dispersal distance; summarized from examples in the literature (after Sanford and Kelly 2011).
Because microgeographic local adaptation is a potential equilibrium result, spatial scale alone does not distinguish these phenomena. As implied by the diagonal gradient in Fig. 1, local adaptation and cohort adaptation likely occur simultaneously in some dispersal/selection scale parameter space. Local adaptation is the component of adaptive differentiation that is balanced with average gene flow over many generations, and cohort adaptation ephemerally contributes microgeographic differentiation in accordance with spatial environmental variation experienced by a cohort.
It may help to describe some unambiguous examples where local adaptation can be completely rejected as a contributing mechanism. Schmidt and Rand (1999) documented cohort adaptation in the barnacle Semibalanus balanoides. A desiccation selection gradient across intertidal zones recurrently generated an allele frequency cline in adult barnacles that was not present in settling juveniles. After a cycle of cohort adaptation, adults just meters apart release offspring with very different genotype frequencies at the mannose-6-phosphate (MPI) locus, only to have them homogenized during months of pelagic larval dispersal such that local adaptation is nearly impossible. For a long time the best examples of cohort adaptation involved single loci of large effect, but that may not be representative of its most pervasive effects. The American eel (Anguila rostrata), for instance, is the quintessential panmictic population because juveniles emerge from a common (presumably well mixed) spawning ground in the Sargasso Sea and migrate indiscriminately to estuaries along the entire eastern seaboard. Over and over again, every life cycle, cohort adaptation differentiates eels in Georgia vs Canada estuaries (macrogeographic scale; Gagnaire, 2012), and in shallow polluted tributaries versus cleaner feeder streams of a single estuary (mesoscale; La Porte, 2016). Cohort adaptation in this case led to polygenic signatures of selection, apparently related to life-history and tolerance phenotypes, respectively.
There are many ways in which cohort adaptation can have demographic and ecological consequences, largely unexplored to date (but see Marshall et al. 2010). Adaptive phenotypic plasticity has limits; where it is insufficient to protect individuals from suboptimal environments cohort adaptation can maintain population persistence or higher local fitness. In this way, cohort adaptation potentially broadens the realized physical niche beyond what plasticity allows. High fecundity and broad dispersal confers a dynamic responsiveness to environmental heterogeneity that likely makes species more resilient to environmental change. Whereas local adaptation and even cohort adaptation can lag behind environmental change in some long lived species like trees (Brown et al. 2019), cohort adaptation in species with shorter generation times can generate more temporally as well as spatially ‘fine tuned’ patterns of local mean fitness. In addition, if mating is local then cohort adaptation leads to genotypically distinct offspring getting produced in different habitat patches so that genetic variance for traits under selection is higher in the overall propagule pool than would be true under a panmictic model. In theory, the higher genetic variance provides a greater capacity for responses to selection by each new metapopulation cohort, increasing mean fitness.
The evolutionary prediction that cohort adaptation maintains balanced polymorphisms has long been appreciated in theory, but remains poorly documented. A recent and fascinating exception is again with the Semibalanus barnacle studied by David Rand’s lab in which they showed (conservatively) that 4% of the transcriptome shows evidence of polymorphisms that have persisted for at least a million years (Nunez et al. 2020), far longer than expected under genetic drift.
On a more practical level, if our understanding of genotypic diversity is shaped primarily by neutral patterns of gene flow and traditional perspectives on adaptation then approaches to broodstock choice for either selective breeding or hatchery-based population supplementation could be suboptimal. For eastern oysters (Crassostrea virginica), subject to both these breeding goals, broodstock collections anywhere in an estuary might be considered genetically exchangeable. In fact, at either end of the estuarine salinity gradient, and perhaps across other environmental transitions as well (subtidal/intertidal; Li et al. 2018), oysters differ at adaptive loci (Eierman and Hare 2015). The Hare Lab is collaboratively investigating the genomic extent and repercussions of cohort adaptation in Delaware Bay oysters in the Selection along Estuarine Gradients in Oysters (SEGO) project, funded by NSF (with co-PIs Daphne Munroe and Elizabeth North).
For a species experiencing cohort adaptation, conventional sampling of adults to test for local adaptation will generate a positive result; for example, average fitness will be higher at ‘home’ than the average fitness measured for the same genotype transplanted to multiple ‘away’ habitats. Thus, for species susceptible to cohort adaptation, additional analyses are required to distinguish cohort adaptation from local adaptation because in many cases these processes will co-occur. What is required to empirically separate these two processes? We want to highlight two potentially useful approaches here. The first, depicted in Fig. 2, predicts that the extent of observable cohort adaptation will depend on the timing of sampling after dispersal and before selection on a cohort is ‘complete’. Sampling of adults to set up common gardens will show adaptive differentiation whereas sampling early post-settlement juveniles (i.e., before much of the cohort selection), and setting up common gardens with those juveniles, is expected to show much less or no differentiation. The difference quantifies cohort adaptation for that cohort. Because the traits under selection will be unknown in many cases, another approach we are using in the SEGO project is to compare whole genome sequences from population samples across an environmental gradient and between survivors and nonsurvivors in experimental challenges. Ideally these genomic comparisons should be made at several life history stages. If the magnitude and genomic prevalence of adaptive differentiation appears similar across the environmental gradient at each life stage then cohort adaptation is rejected and we need to revisit assumptions about the scale of homogenizing larval dispersal, or the acuity of habitat choice.
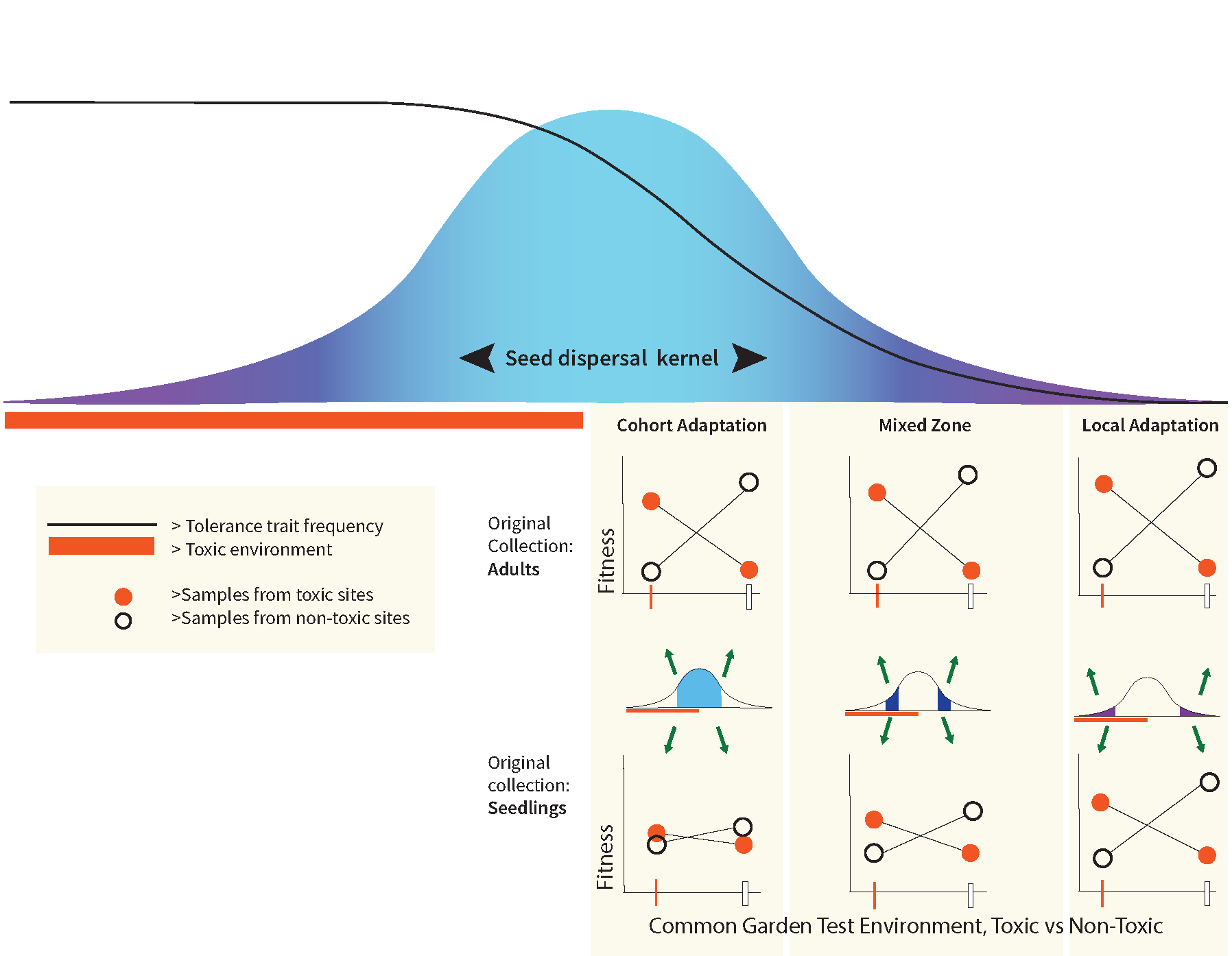
Figure 2: Empirically distinguishing effects of local adaptation and cohort adaptation in the context of a sharp environmental transition relative to average dispersal. In this conceptual diagram the analysis of relative fitness is done with collections at different distances from the environmental transition (varying the level of immigrant seeds from across the transition) and with collections of adults vs. an early life history stage (varying the degree to which cohort selection has acted prior to sampling). The figure depicts results qualitatively expected when cohort selection has a stronger impact near the environmental transition and diminishing effects relative to local adaptation away from the transition. If adults are collected for common gardens, cohort adaptation combined with local adaptation make for strong fitness differences all along the dispersal gradient. Collecting seedlings (before cohort selection) removes cohort adaptation so that fitness differences are attributable to local adaptation with different levels of gene flow constraint.
Darwin’s theory of natural selection was inspired in part by the variation among conspecifics, the raw material of evolution which in turn begets higher forms of diversity. Existing theory and examples have taught us much about the emergent properties of populations at temporal and spatial scales so small that evolutionary biologists often pass them over. More empirical research into the conditions supporting cohort adaptation, and its ecological and evolutionary impact will help us put its effects into context relative to adaptive plasticity and local adaptation. As anthropogenic activities transform the biosphere, reshaping terrestrial and aquatic ecology at a rapid pace, our ability to mobilize the genetic variation existing within and among populations to mitigate this damage depends on how well we understand contemporary population responses to selection.
We would be pleased to hear from you about the ideas in this blog post.
Avi Simon
Junior Biological Sciences major at Cornell University
Matthew Hare
Associate Professor in the Department of Natural Resources and the
Environment, Cornell
mph75[at]cornell.edu
The Hare Lab uses genetics and genomics to investigate evolutionary (and pseudo-evolutionary) processes that could constrain or facilitate population responses to anthropogenic environmental change. These processes include gene flow, adaptation, hybridization and introgression, and population supplementation with domesticated strains.
Literature Cited
Antonovics, J. & Bradshaw, A. D. Evolution in closely adjacent plant populations VIII. Clinal patterns at a mine boundary. Heredity 25, 349–362 (1970).
Browne, L., Wright, J.W., Fitz-Gibbon, S., Gugger, P.F. and Sork, V.L. Adaptational lag to temperature in valley oak (Quercus lobata) can be mitigated by genome-informed assisted gene flow. Proceedings of the National Academy of Sciences, 116(50), pp.25179-25185 (2019).
Bumpus, H.C. The elimination of the unfit as illustrated by the introduced sparrow, Passer domesticus: (a fourth contribution to the study of variation). Biol. Lect. Woods Hole Mar. Biol. Sta. 6: 209-226 (1899).
Burford, M., Scarpa, J., Cook, B. & Hare, M.P. Local adaptation of a marine invertebrate with a high dispersal potential: evidence from a reciprocal transplant experiment of the eastern oyster Crassostrea virginica. Mar. Ecol. Prog. Ser. 505, 161–175 (2014).
Eierman, L.E. and Hare, M.P. Reef specific patterns of gene expression plasticity in eastern oysters (Crassostrea virginica). Journal of Heredity, special issue on phenotypic plasticity and evolution 107:90–100 (2015).
Gagnaire, P.-A., Normandeau, E., Côté, C., Møller Hansen, M. & Bernatchez, L. The genetic consequences of spatially varying selection in the panmictic American eel (Anguilla rostrata). Genetics 190, 725–736 (2012).
Grant, P. R. & Grant, B. R. Predicting microevolutionary responses to directional selection on heritable variation. Evolution 49, 241–251 (1995).
Hays, C. G. Adaptive Phenotypic Differentiation across the intertidal gradient in the alga Silvetia compressa. Ecology 88, 149–157 (2007).
Hoekstra, H. E., Krenz, J. G. & Nachman, M. W. Local adaptation in the rock pocket mouse (Chaetodipus intermedius): natural selection and phylogenetic history of populations. Heredity 94, 217–228 (2005).
Kittelson, P. M. & Maron, J. L. Fine-scale genetically based differentiation of life-history traits in the perennial shrub Lupinus arboreus. Evolution 55, 2429–2438 (2001).
Laporte, M. et al. RAD sequencing reveals within-generation polygenic selection in response to anthropogenic organic and metal contamination in North Atlantic eels. Molecular Ecology 25, 219–237 (2016).
Levene, H. Genetic Equilibrium When More Than One Ecological Niche is Available. The American Naturalist 87, 331–333 (1953).
Levins, R. Evolution in Changing Environments: Some Theoretical Explorations. (Princeton University Press, 1968).
Li, A., Li, L., Wang, W., Song, K. and Zhang, G., 2018. Transcriptomics and fitness data reveal adaptive plasticity of thermal tolerance in oysters inhabiting different tidal zones. Frontiers in physiology, 9, p.825.
McNeilly, T. Evolution in closely adjacent plant populations III. Agrostis tenuis on a small copper mine. Heredity 23, 99–108 (1968).
Nunez, J. C. B. et al. Footprints of natural selection at the mannose-6-phosphate isomerase locus in barnacles. PNAS 117, 5376–5385 (2020).
Rey, C. et al. Within-Generation Polygenic Selection Shapes Fitness-Related Traits across Environments in Juvenile Sea Bream. GENES 11, (2020).
Richardson, J. L., Urban, M. C., Bolnick, D. I. & Skelly, D. K. Microgeographic adaptation and the spatial scale of evolution. Trends Ecol Evol 29, 165–176 (2014).
Sanford, E. & Kelly, M. W. Local adaptation in marine invertebrates. Ann Rev Mar Sci 3, 509–535 (2011).
Schmidt, P. S. & Rand, D. M. Intertidal Microhabitat and Selection at Mpi: Interlocus Contrasts in the Northern Acorn Barnacle, Semibalanus Balanoides. Evolution 53, 135–146 (1999).
Therkildsen, N. O. et al. Contrasting genomic shifts underlie parallel phenotypic evolution in response to fishing. Science 365, 487–490 (2019).
Vendrami, D. L. J. et al. RAD sequencing sheds new light on the genetic structure and local adaptation of European scallops and resolves their demographic histories. Scientific Reports 9, 7455 (2019).
Warner, R. R. Evolutionary ecology: how to reconcile pelagic dispersal with local adaptation. Coral Reefs 16, S115–S120 (1997).
Wittkopp, P. J. et al. Local adaptation for body color in Drosophila americana. Heredity 106, 592–602 (2011).