Recently the IPCC report on the status and trajectory of the global ocean and the cryosphere (the portion of the earth where water mostly exists in a solid state) was released. It paints a clear picture of a rapidly shifting ecosystem that has already undergone remarkable changes in the span of a few decades and is projected to continue changing along similar trajectories (Fig. 1). It’s worth noting that while the report primarily visualizes the difference between the “best case” anthropogenic CO2 reduction scenario RCP 2.6, and the “worst case” business as usual RCP 8.5 scenario, CO2 emissions since 2005 suggest that the RCP 2.6 scenario is already too low to adequately represent future atmospheric CO2 levels (Friedlingstein et al. 2014; Senderson et al. 2016; Millar et al. 2017).
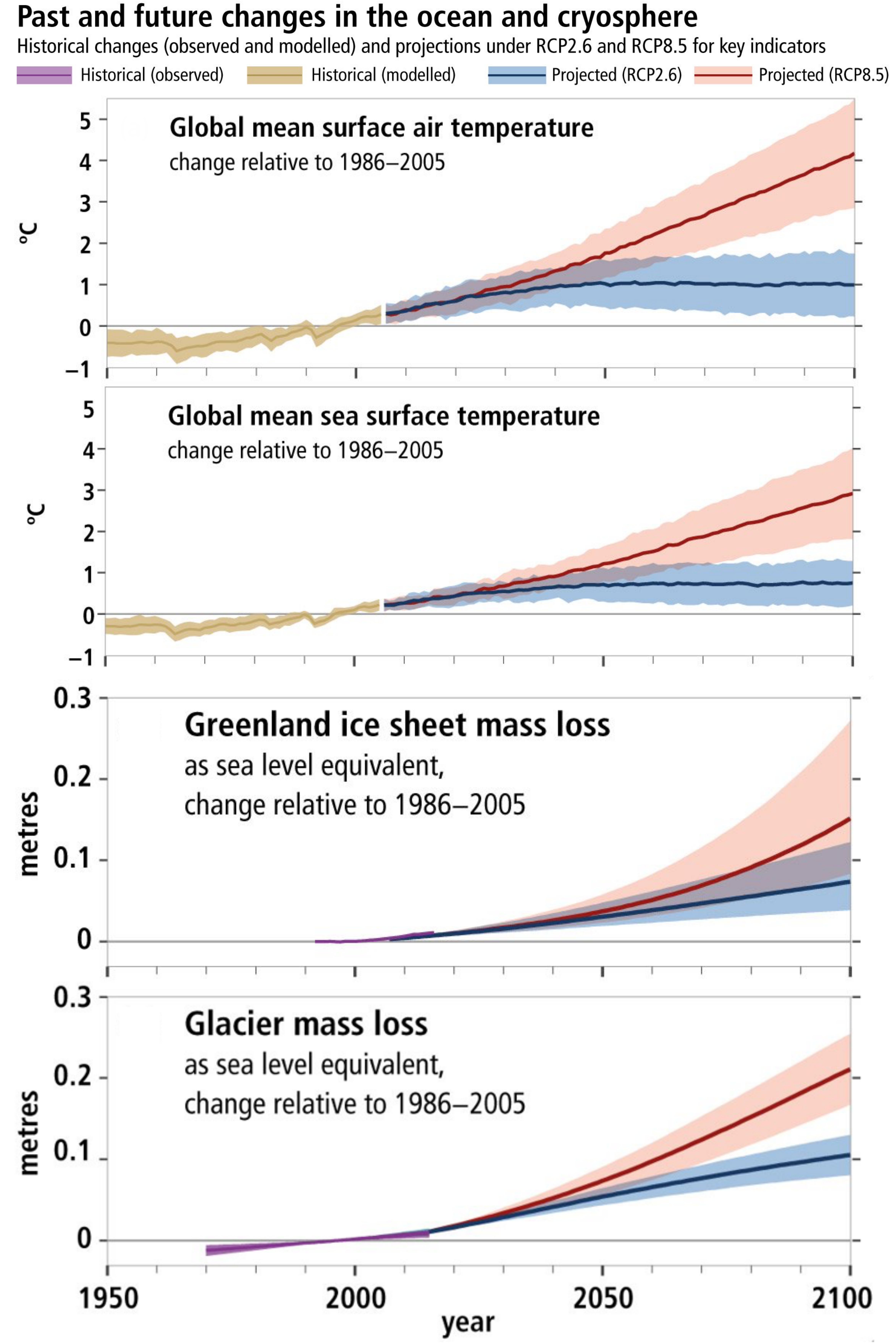
Figure 1. Observed and modeled historical changes in the ocean and cryosphere since 1950. Modified from IPCC (2019) Figure SPM.1.
The report explicitly highlights the important role evolutionary adaptation will play in the ongoing survival of marine populations, citing Scahum and Collins (2014), the report states that while much attention has been given to the evolutionary origin of traits within populations, far fewer studies have considered the stability of trait evolution, or the evolutionary impacts at the level of ecosystems or community function. It goes on the state that while natural adaptation may not be able to keep pace with the rate and magnitude of changes in the ocean and the cryosphere, human assisted evolutionary adaptation may be able to assist in the mitigation of ecosystem decline.
Biologists that are invested in marine population dynamics, especially those populations lacking the resource influence from historical economic investment, i.e. marine invertebrate populations, are then faced with the challenge of predicting evolutionary responses in multiple populations across multiple biological systems across a changing environment with limited molecular resources. Experimental evolution studies such as Pespeni et al. (2013) and Hinners et al. (2017) offer valuable insight into the response potential for extant marine populations, but such projects can be slow to set up, expensive, and prone to complications over a necessarily long-term observable time period.
Even as more and more researhcers gain access to cheaper whole-genome sequencing, the ability to assess evolutionary responses in de novo systems is contingent on interpretations derived from allele frequencies; and as understanding grows of the confluence of effects from genomic architecture, molecular redundancy at the level of an individual up to populations, and signatures of multilocus trait evolution, all in the absence of a broader empirical background information, these signatures can simultaneously tell multiple stories (Poh et al. 2014, Yeaman 2015, Pavlidis & Alachiotis 2017).
One avenue forward in untangling these multiple narratives is the implementation of forward genetic simulations. Multiple software are available that attempt to reconstruct genetic signatures from coalescent based approaches (e.g. CDPOP, SPLATCHES2, and msprime to name just a few), and while coalescent based approaches can operate quickly across many generations of simulations they can be limited in the complexity of the simulated genomic architecture and population demography considered. Forward simulation software like simuPop and SLiM can allow for greater flexibility in this regard, and with the recent implementation of the pyslim Python API, which allows for combining SLiM simulation output with neutral mutations generated by msprime, the tenability of running increasingly complex forward simulations is greater than ever before (Haller et al. 2019b). An example simulation is visualized in Figure 2.
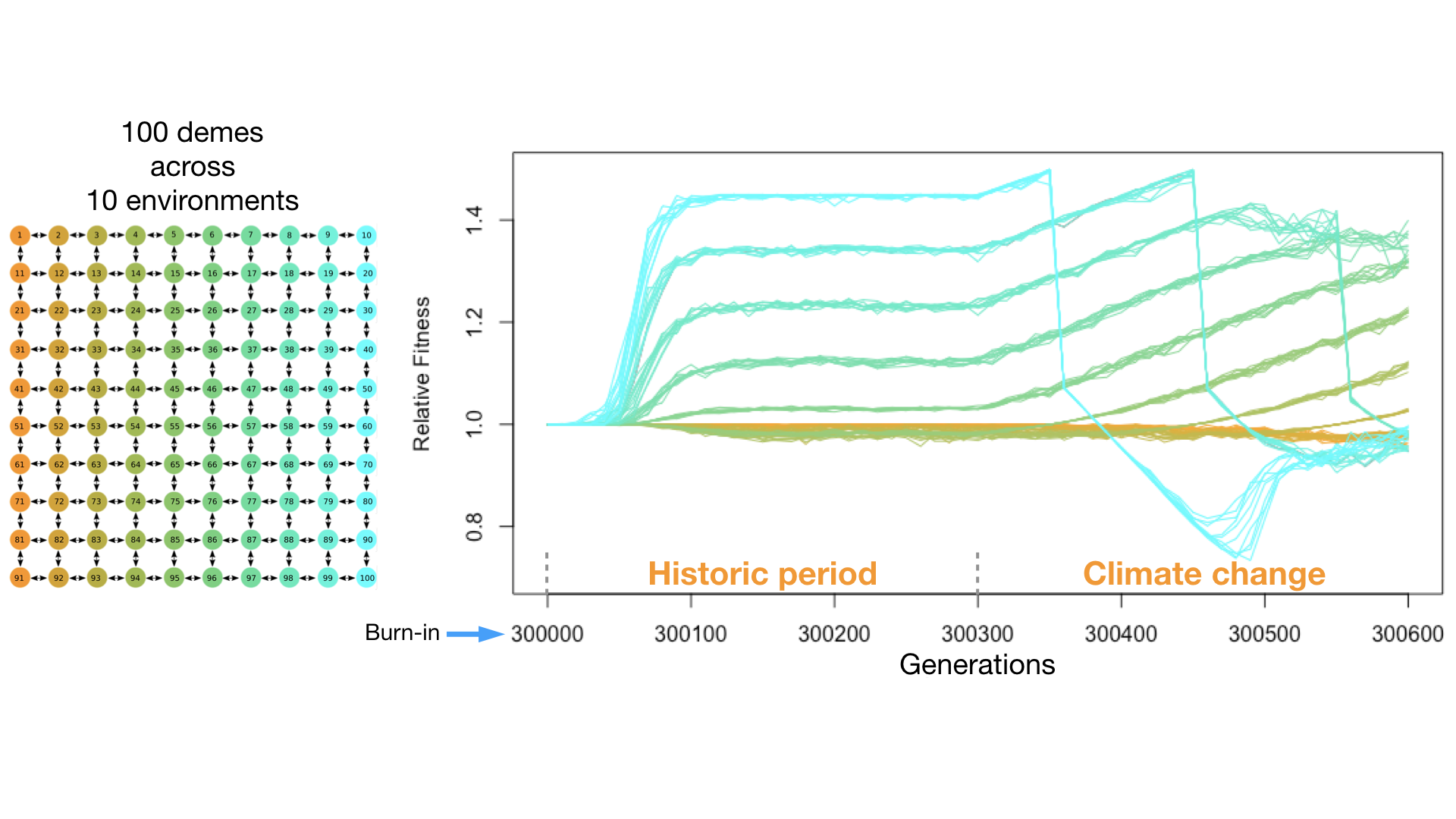 Figure 2. Visualization of an example forward simulation, generated with SLiM 3. An interconnected meta-population consisting of 10,000 individuals arranged across 100 demes, each with a 1Mb diploid genome and 10 recombinant groups. Across the meta-population lies an environmental gradient, represented by the color scheme orange to cyan, which affects the relative fitness of each individual. Over time, the environment is made to shift to represent the effects of climate change, which impacts the pattern of average relative fitness observed within each deme.
Figure 2. Visualization of an example forward simulation, generated with SLiM 3. An interconnected meta-population consisting of 10,000 individuals arranged across 100 demes, each with a 1Mb diploid genome and 10 recombinant groups. Across the meta-population lies an environmental gradient, represented by the color scheme orange to cyan, which affects the relative fitness of each individual. Over time, the environment is made to shift to represent the effects of climate change, which impacts the pattern of average relative fitness observed within each deme.
As highlighted by Palumbi et al. (2019), the potential trade-offs involved in adapting to warmer more acidic marine environments will have unclear affects on long term population dynamics in marine environments. A holistic approach incorporating considerations of both historic and predicted / environmental and genomic data will be necessary moving forward if we are to successfully predict “how human actions may assist evolutionary adaptation and thereby possibly enhance the resilience of natural systems to climate change pressures” (IPCC 2019).
Biography
Áki Jarl Láruson is a Postdoctoral Researcher in the lab of Dr. Katie Lotterhos, in the Department of Marine and Environmental Sciences at Northeastern University. Dr. Láruson studies approaches to detect genomic signatures of adaptation, as well as molecular evolution in marine invertebrates.
Bibliography
Beaugrand, G. & Kirby, R. (2018) How do marine pelagic species respond to climate change? Theories and observations. Annu. Rev. Mar. Sci. 10:169–97. Friedlingstein, P.; Andrew, R.M.; Rogelj, J.; Peters, G.P.; Canadell, J.G.; Knutti, R.; Luderer, G.; Raupach, M.R.; Schaeffer, M.; van Vuuren, D. P.; Le Quéré, C. (2014) Persistent growth of CO2 emissions and implications for reaching climate targets. Nat Geosci 7, 709–715. Gunderson, A.R. & Stillman, J.H. (2015) Plasticity in thermal tolerance has limited potential to buffer ectotherms from global warming. Proc. R. Soc. B. 282(1808). Haller, B.C.; Messer, P.W. (2019a) SLiM 3: Forward genetic simulations beyond the Wright–Fisher model. Mol. Biol. Evol. 36(3):632–637. Haller, B.C.; Galloway, J.; Kelleher, J.; Messer, P.W.; Ralph, P.L. (2019b) Tree‐sequence recording in SLiM opens new horizons for forward‐time simulation of whole genomes. Mol. Ecol. Resour. 19:552– 566. Hinners, J.; Kremp, A.; Hense, I. (2017) Evolution in temperature-dependent phytoplankton traits revealed from a sediment archive: do reaction norms tell the whole story? Proc. R. Soc. B. 284(1888). IPCC (2019) Summary for Policymakers. In: IPCC Special Report on the Ocean and Cryosphere in a Changing Climate [H.-O. Pörtner, D.C. Roberts, V. Masson-Delmotte, P. Zhai, M. Tignor, E. Poloczanska, K. Mintenbeck, M. Nicolai, A. Okem, J. Petzold, B. Rama, N. Weyer (eds.)]. In press. Kelleher, J.; Etheridge, A.M.; McVean, G. (2016) Efficient coalescent simulation and genealogical analysis for large sample sizes, PLoS Comput Biol 12(5): e1004842 Landguth, E.L. and Cushman, S.A. (2010) cdpop: A spatially explicit cost distance population genetics program. Mol. Ecol. Resour. 10:156-161. Millar, R.J.; Fuglestvedt, J.S.; Friedlingstein, P.; Rogelj, J.; Grubb, M.J.; Matthews, H.D.; Skeie, R.B.; Forster, P.M.; Frame, D.J.; Allen, M.R. (2017) Emission budgets and pathways consistent with limiting warming to 1.5 °C. Nat. Geosci. 10, 741–747. Palumbi, S.R.; Evans, T.; Pespeni, M.H.; Somero, G. (2019) Present & future adaptation of marine species assemblages: DNA based insight into climate change from studies of physiology, genomics, and evolution. Oceanography 32(3):83-93. Pavlidis, P. & Alachiotis, N. (2017) A survey of methods and tools to detect recent and strong positive selection. J. Biol Res-Thessalon. 24, 10.1186/s40709-017-0064-0, 28405579 Peng, B. & Kimmel, M. (2005) simuPOP: a forward-time population genetics simulation environment. Bioinformatics 21(18):3686-7. Pespeni, M.H.; Sanford, E.; Gaylord, B.;Hill, T.M.; Hosfelt, J.D.; Jaris, H.K.; LaVigne, M.; Lenz, E.A.; Russell, A.D.; Young, M.K.; Palumbi, S.R. (2013) Evolutionary change during experimental ocean acidification. PNAS 110(17):6,937–6,942 Poh, Y.P.; Domingues, V.S.; Hoekstra, H.E.; Jensen, J.D. (2014) On the prospect of identifying adaptive loci in recently bottlenecked populations. PLOS ONE 9(11): e110579. Ray, N.; Currat, M.; Foll, M.; Excoffier L. (2010) SPLATCHE2: a spatially explicit simulation framework for complex demography, genetic admixture and recombination. Bioinformatics 26(23):2993-4. Sanderson, B. M., O’Neill, B. C. & Tebaldi, C. (2016) What would it take to achieve the Paris temperature targets? Geophys. Res. Lett. 43, 7133–7142. Schaum, C.E. & Collins, S. (2014) Plasticity predicts evolution in a marine alga. Proc. R. Soc. B. 281(1793). Yeaman, S. (2015) Local adaptation by alleles of small effect. Am Nat 186(S1):S74-S89